scDALI (single-cell differential allelic imbalance) is a statistical model and analysis framework that leverages allele-specific analyses of single-cell data to decode cell-state-specific genetic regulation.
The key idea of the scDALI workflow is the integration of two independent signals that can be extracted from the same single cell sequencing experiment: total counts and allele-specific quantifications. First, total read counts are used to define a cell state representation, applying established methods for dimensionality reduction, cell clustering or the inference of pseudo-temporal orderings. Second, from the same dataset, allele-specific counts from matched cells are extracted, which allow for quantifying allelic imbalances and therefore genetic effects. scDALI connects these two signals in order to detect heterogeneous (cell-state-specific) as well as homogeneous imbalances.
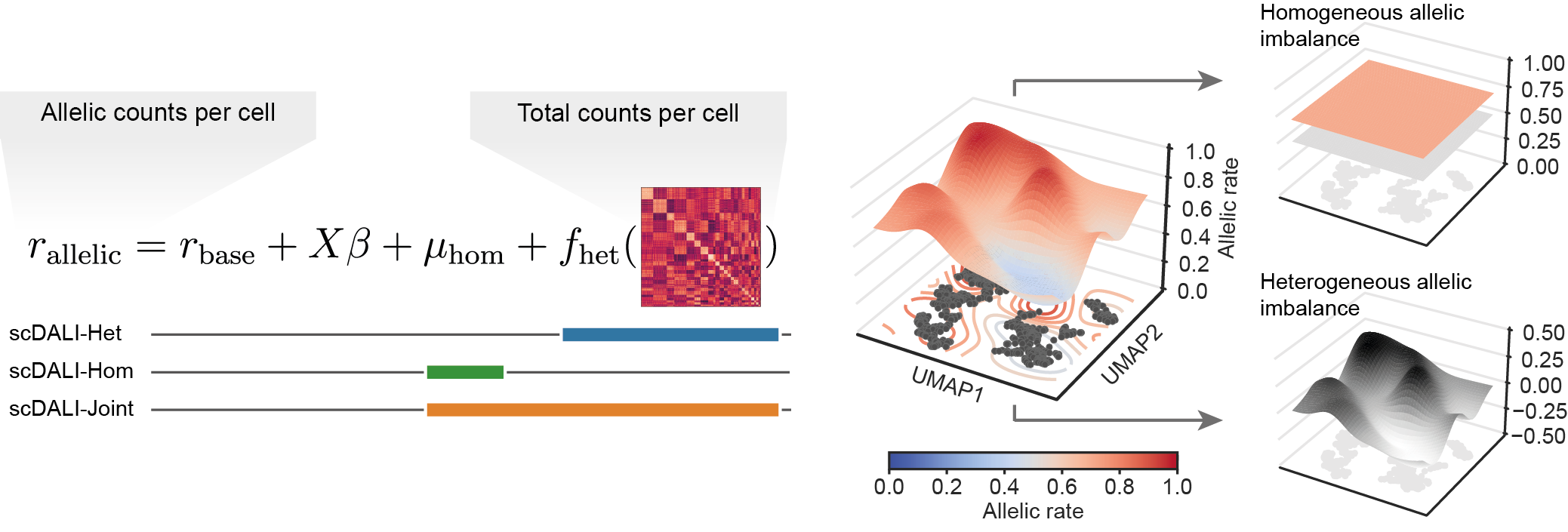
Conceptionally, the scDALI approach is similar to differential gene expression testing. However, instead of identifying differentially expressed genes between cell types or states, scDALI captures differential allelic imbalance. scDALI can model both continuous and discrete cell states and does not require to discretize the cell-state space a priori.
For more information on the scDALI method along with evaluations on real and simulated data, please have a look at our preprint!